|
BioSyM Research
Thrust 1: Molecular Scale Tools
 |
|
|
|
|
Three highly stretched DNA structures |
Multispectral force fluorescence microscope |
Cytoskeletal rearrangement in presence of a molecularly crowded cellular environment |
Design of Resistance-Avoiding Hepatitis-C Inhibitor |
BioSyM’s molecular manipulation tools include a suite of capabilities that are focused on high-resolution measurement of how and why biomacromolecules interact under crowded, confined, and mechanically strained conditions common in biological systems. Our force probe arsenal includes custom-built magnetic, optical, and atomic force microscopes, many of which have been integrated within high-end multi-dimensional imaging platforms capable of simultaneous measurement of force, color, and time. Specific projects include nanofluidic studies of DNA structure and stability; rheology of biogels as related to health and disease; novel approaches to drug target identification through computer-modeling enabled analysis of biological pathways; biological-part based synthetic materials, structures, sensors, and machinery. These studies will advance our understanding of the foundational principles of molecular interactions and bio-systems integration.
Capabilities Developed:
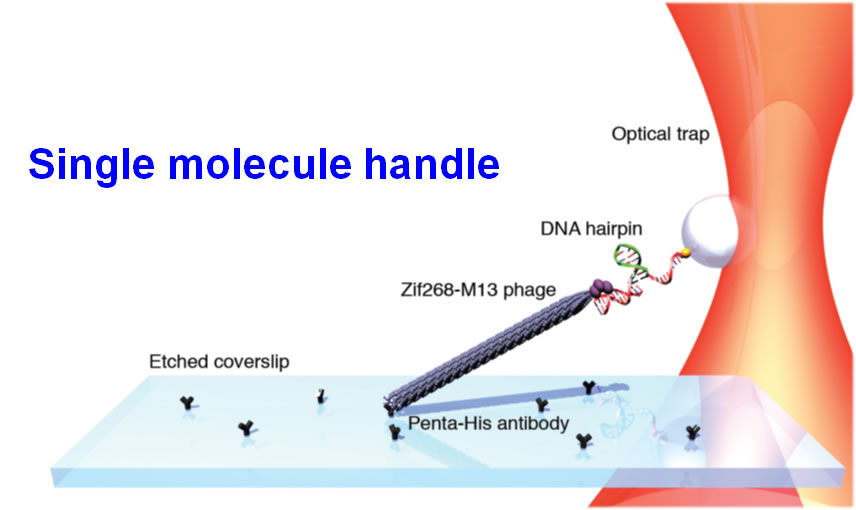
- Combined trap and single molecule fluorescence microscope: This instrument has the ability to measure protein unfolding, DNA stretching, motor protein motility and single cell mechanical studies. It features an optical trap with multiple color single molecule fluorescence excitation (Blue, green, red). The optical (double) trap is able to apply forces above 100 pN and features position resolution of a single nm.
- Pump-Probe Thermal Imaging Microscope: This instrument is capable of scanning a surface and precisely locating small gold nano particles that are resonant with the pump laser. Instrument resolution is close or below so called diffraction resolution.
- Recombinant Protein Production and Purification: Our lab has developed the infrastructure to make mutations to single proteins and produce them recombinantly using standard methods such as e-coli based protein production. We are generating model 4 helix bundle structures as well as two different types of motor protein constructs.
- DNA Origami: Our lab has also adapted methods for production of DNA-orgami smart scaffolds. Here we are using the DNA template as a geometry organizer, mixing these structures with coupled gold nano particle plasmonics.
- Video particle tracking technique: This allows high speed imaging of particles in a biological system and allows investigation of the mechanical properties of Biofilms and complex fluids like DNA, protein etc.
- Manipulation of DNA by magnetic field (magnetic tweezers) and Electric field: Provides insights of the molecular structure of DNA and its dynamics.
- Computational models and algorithms for DNA mechanics: To simulate the conformations and dynamics of biopolymers, e.g. DNA, under various conditions, such as confinement by microfluidic channels, and forces by external fields. The simulation provides molecular details and quantitative results to understand underlying mechanisms that controls DNA in microfluidics, and helps the practical applications, such as genome mapping by microfluidics and DNA sequencing by nanopores.
People
|
Research Highlights
|
|
Binu Kundukad (SMART), Piewen Cong (NUS), Johan R. C. van der Maarel (NUS), and Patrick S. Doyle (MIT/SMART)
HU is a protein that plays a role in various bacterial processes including compaction, transcription and
replication of the genome. Here, we use atomic force microscopy to study the effect of HU on the stiffness and supercoiling of double-stranded DNA. First, we measured the persistence length, height profile, contour length and bending angle distribution of the DNA–HU complex after different incubation times of HU with linear DNA. We found that the persistence and contour length depend on the incubation time. At high concentrations of HU, DNA molecules first become stiff with a larger value of the persistence length. The persistence length then decreases over time and the molecules regain the flexibility of bare DNA after 2 h. Concurrently, the contour length shows a slight increase. Second, we measured the change in topology of closed circular relaxed DNA following binding of HU. Here, we observed that HU induces supercoiling over a similar time span as the measured change in persistence length. Our observations can be rationalized in terms of the formation of a nucleoprotein filament followed by a structural rearrangement of the bound HU on DNA. The rearrangement results in a change in topology, an increase in bending flexibility and an increase in contour length through a decrease in helical pitch of the duplex.
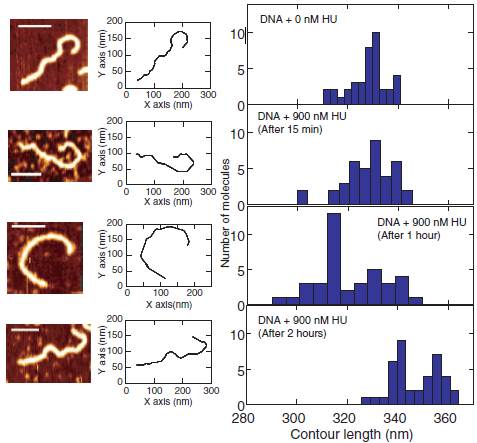
Representative AFM image, corresponding contour and contour length distribution for bare DNA and incubated with 900nM HU for 15 min, 1 h and 2 h from top to bottom, respectively. The scale bars denote 100 nm. The distributions are obtained from a pool of ~40 molecules each.
Yun Soo Kim (SMART), Binu Kundukad (SMART) Abdollah Allahverdi (NTU), Lars Nordenskold (NTU), Patrick S. Doyle (MIT/SMART) and Johan R. C. van der Maarel (NUS)
Topoisomerase II (TOP2) regulates the topology of DNA by catalysis of a double strand passage reaction. Inhibition of this reaction prevents cell replication, and, thus, is a pathway targeted by anticancer drugs. Some details regarding the cell-killing mechanism are unknown and assays to screen for anticancer drugs are not well established. Here, we have studied the gelation of linear and circular DNA using microrheology assays.
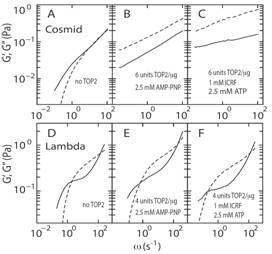 |
Circular DNA (Cosmid) forms a gel when incubated with AMPPNP and ICRF where as linear DNA (lambda) does not form a gel under similar conditions |
3. Macromolecular crowding effects on in vitro cell culture systems
Adam S. Zeiger (MIT/SMART), Felicia C. Loe (SMART), Ran Li (MIT), Michael Raghunath (NUS), and Krystyn J. Van Vliet (MIT/SMART)
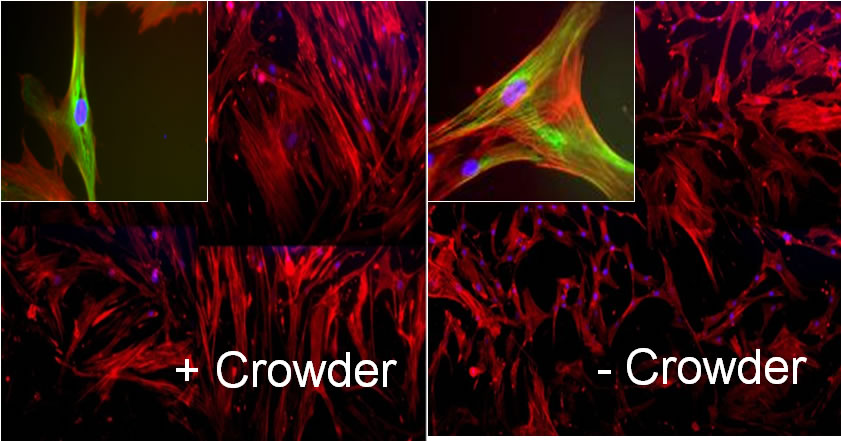
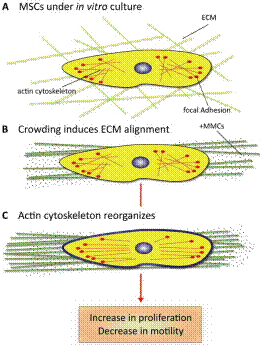
A crucial part of advanced in vitro platform design is increasing fidelity to physiological environments when necessary. For example, contemporary in vitro culture systems lack the level of macromolecular concentrations that is found in vivo, and thus poorly represent those conditions. We have induced macromolecular crowding in vitro using synthetic macromolecular globules of nm-scale radius at in vivo levels of fractional volume occupancy. We quantified the impact of induced crowding on the extracellular and intracellular protein organization of human mesenchymal stem cells (MSCs) via immunocytochemistry, atomic force microscopy (AFM), and AFM-enabled nano-indentation. Macromolecular crowding in extracellular culture media is found to directly induce supramolecular assembly and alignment of extracellular matrix proteins deposited by cells, which in turn increases alignment of the actin cytoskeleton. The resulting cell-matrix reciprocity further affects adhesion, proliferation, and migration behaviour of MSCs. Macromolecular crowding can thus aid the design of more physiologically relevant in vitro studies and devices for MSCs and other cells, by increasing the fidelity between materials synthesized by cells in vivo and in vitro. Thus, this project within Thrust 1 has immediate implications in Thrust 2, Multifunctional Cytometry.
|
Schematic of cell and matrix reciprocity under macromolecular crowding. (A) Adherent mesenchymal stromal cell (MSC) under typical in vitro culture conditions. (B) Upon the addition of macromolecular crowders, excluded volume effects promote bundling and alignment of extracellular matrix (ECM) fibers. (C) In response to the aligned ECM, the intracellular actin cytoskeleton reorganizes to align with the ECM. In addition, the cell secretes more matrix proteins, which are deposited to and further enhance the alignment of the ECM. We find that these reciprocal changes in the matrix material and intracellular cytoskeletal organization correlate with an increase in proliferation and a decrease in motility of MSCs. |
|
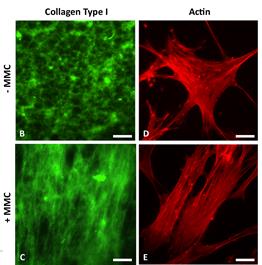
First direct in vitro observation that nanoscale crowding molecules induce alignment of and deposition of extracellular matrix filaments |
|
4. Conformation and Dynamics of DNA in Nanofluidic Devices
Dai Liang (SMART), Jeremy Jones (MIT), Benjamin Renner (MIT), Patrick Doyle (MIT/SMART), Jie Yan (NUS) and Johan van der Maarel (NUS)
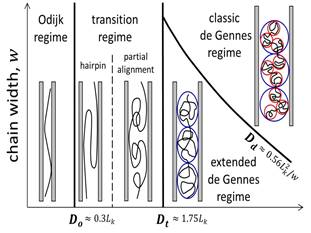
We have studied conformation and dynamics of DNA in nanofluidic devices. The confinement can be applied to stretch DNA, modify DNA topology, slow down DNA dynamics, and alter the responses to external fields such as flow and electric fields. The major methodologies are Monte-Carlo simulation and scaling theory. The results provide new insights to the transition regime from weak to strong confinement. We find that the confinement effect on knotting probability can be monotonic or non-monotonic, depending on the characteristic lengths, including DNA contour length, persistence length and the effective chain width (ionic strength dependent). In addition to well-known similarity in classic de Gennes/Pincus regime, we reveal the subtle difference in the extended de Gennes/Pincus regime. Overall, our research deepens the understanding of DNA behaviors in confinement, especially in the transition regime (channel width ~ 100 nm), which should benefit the practical applications of micro-/nano- fluidic devices to DNA as well as other biopolymers.
|
Schematic illustration of different regimes experienced by a long polymer in a tubular channel when varying the channel cross-sectional dimension and the chain width. D corresponds to diameter for a channel with circular cross section and width or height for a square channel. The black curves represent polymer chains, red circles represent thermal blobs, and blue circles represent the self-avoiding blobs. The boundaries between scaling regimes, Dt and Dd, are determined by our simulations presented in this work. |
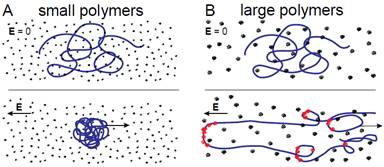
5. Compression and self-entanglement of single DNA molecules under electric field
Ning Du (SMART), Jing Tang (MIT), Jeremy J. Jones (MIT), Ben Renner (MIT), Patrick Doyle (MIT/SMART), Jie Yan (NUS) and Johan van der Maarel (NUS)
Electric fields have been widely used to transport and manipulate DNA in lab-on-a-chip devices with applications in molecular genetics, nucleic acid-based diagnostics, and fundamental studies of polyelectrolytes. We have experimentally demonstrated that addition of small charge-neutral flexible polymers to an ionic buffer can promote compression of single DNA under a uniform electric field. This phenomenon lies in stark contrast with widely observed extension of DNA during capillary electrophoresis in dilute solutions of larger charge-neutral polymers. We propose that this dissimilarity arises from differences in the magnitude of tension created by DNA-polymer collisions, controlled by the properties of the solute polymer. For solutions of small (Mw ≤ 410k) dextran, we show that their ability to promote the compression of single DNA is consistent with a mechanism proposed for aggregation in concentrated macro-ion dispersions by showing collapse of the experimental data. With increasing importance of micro/nanofluidic devices in single molecule research, it is necessary to effectively manipulate DNA conformations. By tuning the solvent of DNA via addition of polymers of varying size and rigidity, one can responsively control DNA configurations via the application of small to moderate electric fields. These insights shed light on the fundamental behavior of polyelectrolytes and provide guidelines for working with the electro-kinetically controlled micro/nanofluidic systems.
|
Schematic of DNA electrophoresing in dilute solutions of polymers. Top: DNA at rest prior to application of electric field. Bottom: Steady state configurations long after switching on the electric field. (A) DNA with “small” polymers (B) DNA with “large” polymers Polymers entrained with DNA contour is shown in red. |
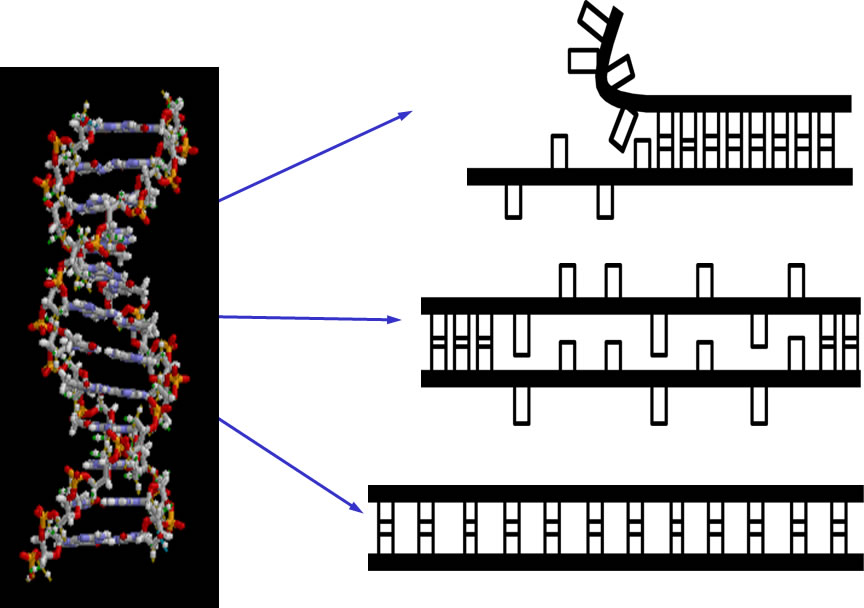
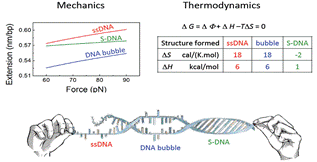
6. The structures and functions of DNA under tension
Xinghua Zhang (SMART), Peiwen Cong (NUS), Patrick Doyle (MIT/SMART), Jie Yan (NUS) and Johan van der Maarel (NUS)
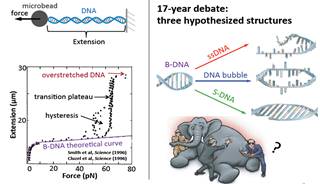
We have developed magnetic tweezers that are capable of grabbing hold of and pulling single DNA molecules. Importantly, we have used our apparatus to resolve a decades-old debate regarding the tension-induced structural transition of DNA into so called S-DNA at large applied forces. Specifically, we found that the DNA overstretching transition through the “B-to-S” transition pathway leads to a highly ordered double-stranded structure without melting of DNA base pairs. The entropy and enthalpy changes during the force-induced internal melting transition of a DNA without nicks or free ends were determined for the first time. We also found that an internal melting transition can happen under physiological conditions. Our results raise interesting questions regarding the exact structure of S-DNA and whether the highly ordered S-DNA structure provides a structural motif that can be used by proteins to bind in vivo. The mechano-thermal stability and inter conversion between these DNA structures have been fully characterized. We plan to explore the potential applications of these transitions in designing novel DNA based mechanical nano-devices.
|
DNA overstretching transitions |
|
All three transitions were identified and characterized |
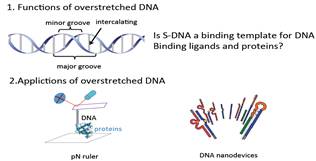
|
Potential functions and applications |
7. Multispectral Force-Fluorescence Microscope
Siew-Kit Hoi (SMART), Taishi Zhang (NUS), Zhenping Guan (NUS), Matt Lang (MIT/SMART), Patrick Doyle (MIT/SMART), Yan Jie (NUS) and Qing-Hua Xu (NUS)
We have developed an integrated fluorescence and force (through: optical, fluidic, magnetic) microscope appropriate for single molecule and cell studies. Custom machining and general assembly of microscope were completed at MIT, and the entire system transferred and commissioned at BioSyM SMART' opto-mechanical suite in CREATE. The instrumentaion features double trap geometry. The combination of the optical tweezers microscopy and single molecule fluorescence spectroscopy has provided the capability for simultaneous manipulation and monitoring of molecular structure in real time. The optical traps can be employed to supply controlled external loads while fluorescent molecules report the concurrent information about macromolecular structure.
|
Multispectral force-fluorescence microscope at SMART-BioSyM
|
8. Understanding Molecular Basis of Mechanotransduction using Computer Simulations
At the molecular level, force induced conformational change can alter protein activity; one possible mechanism for altering activity is through changes in chemical properties such as binding affinity and enzyme activity. In this framework, force-induced alteration of binding affinity, as but one example, would depend on characteristics of the applied force, including direction, magnitude, and point of application. Currently the property space of how applied force affects chemical properties like binding affinity is poorly understood due to the lack of effective experimental data. Here we develop a computational approach and analysis for gaining insight into force modulated binding properties.
The main goal in this project is to understand the molecular basis of mechanotransduction. For this we apply free energy simulation to study the binding response to applied force of focal adhesion targeting domain of the focal adhesion kinase bound to the paxillin motif. Applied force on the protein in different directions helps understand how its direction and magnitude can affect structure-function relationship in biologically relevant systems.
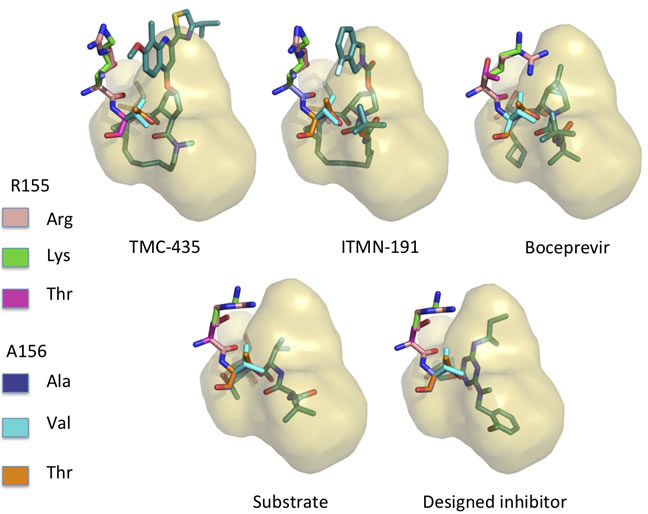
9. Robust inhibition of Hepatitis C viral propagation
The goal is to design mutation resistant inhibitors for hepatitis C virus (HCV) protease using structure-based computational modeling applying inverse design methods to implement the substrate envelope hypothesis. The protease structure was prepared for modeling and design studies using standard approaches. The other important residues that constituted the active substrate to be cleaved were also successfully modeled and substrate envelope is created. According to the synthesis requirement, the scaffold and building blocks are prepared for the design.
Mechanical characteristics of single biological cells are used to identify and possibly leverage interesting differences among cells or cell populations. Fluidity—hysteresivity normalized to the extremes of an elastic solid or a viscous liquid—can be extracted from, and compared among, multiple rheological measurements of cells: creep compliance vs. time, complex modulus vs. frequency, and phase lag vs. frequency. With multiple strategies available for acquisition of this nondimensional property, fluidity may serve as a useful and robust parameter for distinguishing cell populations, and for understanding the physical origins of deformability in soft matter. Here, for three disparate eukaryotic cell types deformed in the suspended state via optical stretching, we examine the dependence of fluidity on chemical and environmental influences around a time scale of 1 s. We find that fluidity estimates are consistent in the time and the frequency domains under a structural damping (power-law or fractional derivative) model, but not under an equivalent-complexity lumped component (spring-dashpot) model; the latter predicts spurious time constants. Although fluidity is suppressed by chemical crosslinking, we find that adenosine triphosphate (ATP) depletion in the cell does not measurably alter the parameter, and thus conclude that active ATP-driven events are not a crucial enabler of fluidity during linear viscoelastic deformation of a suspended cell. Finally, by using the capacity of optical stretching to produce near-instantaneous increases in cell temperature, we establish that fluidity increases with temperature—now measured in a fully suspended, sortable cell without the complicating factor of cell-substratum adhesion.
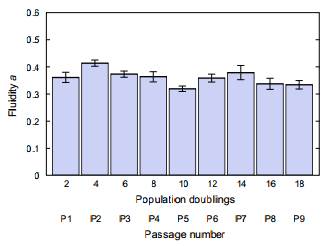
Fluidity is not detectably altered by extended in vitro culturing of primary hMSCs.
|
Journal Publications
|
|
Conference Presentations / Publications
|
- Durgarao Guttela (SMART/MBI), M.Yao (MBI), K.Baker (Univ.of Kent, UK), L.Yang, B.T.Goult (MIT), Patrick Doyle (MIT/SMART) and Jie Yan (NUS), "Calcium-mediated Protein Folding and Stabilization of Salmonella Biofilm-associated Protein A", Journal of Molecular Biology, Jan (2019), pp. 433-443
- Dawei Ding (SMART), Binu Kundukad (SMART), Ambika Somasundar (NUS), Sindhu Vijayan (SUTD), Saif A.Khan (NUS), Patrick Doyle (MIT/SMART), "Design of Mucoadhesive PLGA Microparticles for Ocular Drug Delivery", ACS Applied Bio Materials, Publication Date (Web): August 16, 2018
- Dai Liang (SMART), and Patrick Doyle (MIT/SMART), "Universal Knowt Spectra for Confined Polymers", Macromolecules, 51(16), pp 6327-6333, 2018
- Liang Dai (SMART), Jeremy J. Jones (MIT), Alexander R. Klotz (MIT), Stephen Levy (Binghamton University), and Patrick S. Doyle (MIT/SMART), "Nanoconfinement greatly speeds up the nucleation and the annealing in single-DNA collapse", Soft matter, Published Online Aug 2017.
- Weijie Chi (SUTD), Wenting Yin (CAS), Qingkai Qi (CAS), Qinglong Qiao (CAS), Yuyan Lin (Raffles Institution), Zhuohui Zhu (Raffles Institution), Sindhu Vijayan (SUTD), Michinao Hashimoto (SUTD) Gayathri Udayakumar (Global Indian Intl School), Zhaochao Xu (CAS), Xiaogang Liu (SMART), “Ground-State Conformers Enable Bright Single-Fluorophore Ratiometric Thermometers with Positive Temperature Coefficients”, Materials Chemistry Frontiers, 2017, DOI: 10.1039/C7QM00345E, published online, Aug 2017
- Binu Kundukad**(SMART), Megan Schussman (MIT), Kaiyuan Yang (NUS), Thomas William Seviour**(SCELSE), Liang Yang (SCELSE), Scott A. Rice (SCELSE), Staffan Kjelleberg (SCELSE) and Patrick Doyle (MIT/SMART), “Mechanistic action of weak acid drugs on biofilms” Scientific Reports, 7; 4783 (2017)
- Liu Xiaogang (SMART), J.M.Cole (Cambridge) and Z.Xu (CAS), "Substantial Intramolecular Charge Transfer Induces Long Emission Wavelengths and Mega Stokes Shifts in 6-Aminocoumarins", Journal of Physical Chemistry, 1 June, 2017, Published Online
- L. Zhou, Q. Wang, Y. Tan, M. J. Lang (SMART/Vanderbilt), H. Sun, X. Liu (SMART), "“Rational Development of Near-Infrared Fluorophores with Large Stokes Shifts, Bright One-Photon and Two-Photon Emissions for Bioimaging and Biosensing Applications”, Chemistry - A European Journal, online: 8 May 2017
- Dai Liang (SMART), and Patrick Doyle (MIT/SMART), "Trapping a Knot into Tight Conformations by Intra-Chain Repulsions", Polymers, 9, 57 (2017)
- Dai Liang (SMART), and Patrick Doyle (MIT/SMART), "Effects of Intrachain Interactions on the Knot Size of a Polymer", Macromolecules, Publication online: September 22, 2016
- X. Liu (SMART), Q. Qiao, W. Tian, W. Liu, J. Chen, M. J. Lang (Vanderbilt/SMART), Z. Xu, “Aziridinyl fluorophores demonstrate bright fluorescence and superior photostability through effectively inhibiting twisted intramolecular charge transfer”, Journal of American Chemical Society, 2016, DOI: 10.1021/jacs.6b03924.
- Binu Kundukad (SMART), Thomas Seviour (SCELSE, NTU), Yang Liang (SCELSE, NTU), Scott A. Rice (SCELSE, NTU), Staffan Kjelleberg (SCELSE, NTU), and Patrick S. Doyle (MIT/SMART), "Mechanical properties of the superficial biofilm layer determine the architecture of biofilm", Soft Matter; DOI: 10.1039/C6SM00687F (2016)
- S. C. Chew (SCELSE, NTU), B. Kundukad (SMART), W. K. Teh (SCELSE, NTU), P. Doyle (MIT/SMART) L. Yang (SCELSE, NTU), S. A. Rice (SCELSE, NTU) and S. Kjelleberg (SCELSE, NTU), "Mechanical signatures of microbial biofilms in micropillar-embedded growth chambers", Soft Matter, DOI: 10.1039/c5sm02755a, 2016
- Liang Dai (SMART), C. Benjamin Renner (MIT), Patrick S. Doyle (MIT/SMART), "The polymer physics of single DNA confined in nanochannels", Advances in Colloid and Interface Science 232 (2016) 80–100
- Sonia Brady (Vanderbilt), Sreelatha Sarangapani (SMART), Yinnian Feng (Vanderbilt), Shishir P.S. Chundawat (Rutgers), Matthew J. Lang (Vanderbilt/SMART), "Cellobiohydrolase 1 from Trichoderma reesei degrades cellulose in single cellobiose steps", Nature Communications, 6:10149 (2015)| DOI: 10.1038/ncomms10149
- Liang Dai (SMART), C. Benjamin Renner (MIT), Jie Yan (NUS) Patrick S. Doyle (MIT/SMART), "Coil-globule transition of a single semiflexible chain in slitlikeconfinement", Scientific Reports, 5:18438 (2015) DOI: 10.1038/srep18438
- Peiwen Cong (NUS), Liang Dai (SMART), Hu Chen (NUS), Johan R. C. van der Maarel (NUS), Patrick S. Doyle (MIT/SMART) and Jie Yan (NUS), "Revisit the anomalous bending elasticity of sharply bent DNA", Biophysical Journal (Published online, November 2015)
- Taishi Zhang (NUS/SMART), Nengyue Gao (NUS), Shuang Li (NUS), Matthew J. Lang (Vanderbilt/SMART), and Qing-Hua Xu (NUS), "Single-Particle Spectroscopic Study on Fluorescence Enhancement by Plasmon Coupled Gold Nanorod Dimers Assembled on DNA Origami", J. Phys. Chem. Lett., 2015, 6, 2043–2049
- Liang Dai (SMART), C.Benjamin Renner (MIT) and Patrick Doyle (MIT/SMART), "Metastable Knots in Confined Semiflexible Chains", Macromolecules, (published online in April 2015)
- Liang Dai (SMART), C. Benjamin Renner (MIT), and Patrick S. Doyle (MIT), "Origin of Metastable Knots in Single Flexible Chains", Physical Review Letters, 114, 037801 (2015)
- Binu Kundukad (SMART), Jie Yan (NUS/SMART) and Patrick Doyle (MIT/SMART), "Effect of YOYO-1 on the mechanical properties of DNA", Soft Matter, Published Online 28 Oct 2014
- Xinghua Zhang (SMART), Yuanyuan Qu (NUS), Hu Chen (MBI/NUS), Ioulia Rouzina (Univ. of Minnesota), Shengli Zhang (NUS), Patrick S. Doyl (MIT/SMART),
and Jie Yan (NUS/SMART), Interconversion between Three Overstretched DNA Structures, Journal of American Chemical Society, 136 (45), pp 16073–16080, 2014
- Shimin Le (MBI/NUS), Hu Chen (MBI/NUS), Xinghua Zhang (SMART), Jin Chen (MBI/NUS), K. Neelakanteshwar Patil (IISC, India),
Kalappa Muniyappa (IISC, India), and Jie Yan (NUS/SMART), "Mechanical force antagonizes the inhibitory effects of RecX on RecA filament formation in Mycobacterium
tuberculosis", Nucleic Acids Research, doi: 10.1093/nar/gku899, Published Online on 7th Oct 2014
- Dai Liang (SMART), Benjamin Renner (MIT), Patrick Doyle, "Metastable Tight Knots in Semiflexible Chains", Macromolecules, Published Online,
August 28, 2014
- Su Chuen Chew (SCELSE/NTU), Binu Kundukad (SMART), Thomas William Seviour (SCELSE), Johan R C van der Maarel (NUS), Liang Yang (SCELSE), Scott A. Rice (SCELSE), Patrick Doyle (MIT/SMART) and Staffan Kjelleberg (SCELSE), “Dynamic remodeling of microbial biofilms by functionally distinct exopolysaccharides", mBio, 5 (4), July/August 2014
- Dai Liang (SMART), Johan van der Maarel (NUS) and Patrick Doyle (MIT/SMART), "Extended de Gennes Regime of DNA Confined in a Nanochannel" Macromolecules, Published on web, Mar 25, 2014, DOI:10.1021/ma500326w
- Matthew J. Whitfield (MIT), Wong Cheng J. Lee * (NUS), Krystyn J. Van Vliet (MIT/SMART), “Onset of heterogeneity in culture-expanded bone marrow stromal cells” Stem Cell Research, 11(3), 1365, 2013
- Zhang, C. (NUS), Hernandez-Garcia, A., Jiang, K., Gong, Z., Guttula, D., Ng, S.Y., Malar, P.P., van Kan, J.A., Dai, L.(SMART), Doyle, P.S.(MIT/SMART), de Vries, R., and an der Maarel, J.R.C.(NUS), "Amplified Stretch of Bottlebrush-Coated DNA in Nanofluidic Channels", Nucleic Acids Res., DOI:10.1093/nar/gkt783, 2013.
- Zhang, C.(NUS), Guttula, D., Liu, F., Malar, P.P., Ng, S.Y., Dai, L(SMART)., Doyle, P.S.(MIT/SMART), van Kan, J.A., and van der Maarel, J.R.C.(NUS/SMART), "Effect of H-NS on the elongation and compaction of single DNA molecules in a nanospace", Soft Matter, DOI:10.1039/C3SM51214B, 2013
- Binu Kundukad (SMART), Piewen Cong (NUS), Johan R. C. van der Maarel (NUS), and Patrick S. Doyle (MIT/SMART). “Time-dependent bending rigidity and helical twist of DNA by rearrangement of bound HU protein” Nucleic Acids Research (2013) doi: 10.1093/nar/gkt593
- Dai, L. (SMART), and Doyle, P.S. (MIT/SMART), "Comparisons of a polymer in confinement versus applied force", Macromolecules, DOI: 10.1021/ma400674q (Web) July 29, 2013
- Maloney, J.M (MIT/SMART), Lehnhardt, E., Long, A.F., and Van Vliet, K.J. (MIT/SMART), “Mechanical fluidity of fully suspended biological cells,” Biophysical Journal, 2013, in press
- Cai P (Hokkaido University), Mizutani Y (Hokkaido University), Tsuchiya M (Hokkaido University), Maloney JM (MIT), Fabry B, Van Vliet KJ (MIT/SMART), Okajima T, "Quantifying cell-to-cell variation in power-law rheology," Biophysical Journal, (In press).
- Liang Dai (SMART), Douglas R. Tree, Johan R.C. van der Maarel (NUS), Kevin D. Dorfman, and Patrick S. Doyle (MIT/SMART), "Revisiting blob theory for DNA diffusivity in slitlike confinement", Physical Review Letters, 110, 168105 (2013)
- Zhang Xinghua (SMART), Chen H (MBI), Le S (MBI), Rouzina I (Univ. Minnesota, USA), Doyle P.S. (MIT/SMART), and Yan J (NUS/MBI)., "Revealing the Competition between Peeled-ssDNA, Melting Bubbles and S-DNA during DNA Overstretching by Single-Molecule Calorimetry", Proc. Natl. Acad. Sci., published online (Feb 2013)
- Yun Soo Kim (SMART), Binu Kundukad (SMART) Abdollah Allahverdi (NTU), Lars Nordenskold (NTU), Patrick S. Doyle (MIT/SMART) and Johan R. C. van der Maarel (NUS), “Gelation of the genome by topoisomerase II targeting anticancer agents”, Soft Matter, Advance Article Published online, DOI: 10.1039/C2SM27229F (2013)
- Dai, Liang (SMART), Ng, Siow; Doyle, Patrick (MIT/SMART); Van Der Maarel, Johan (NUS), "Conformation model of back-folding and looping of a single DNA molecule confined inside a nanochannel", ACS Macro Letters, 1, 1046 (2012)
- Liang Dai (SMART), Johan R. C. van der Maarel (NUS), and Patrick S. Doyle (MIT/SMART), "Effect of Nanoslit Confinement on the Knotting Probability of Circular DNA", ACS Macro Letters, 1, 732−736 (2012)
- Adam S. Zeiger (MIT/SMART), Benjamin Hinton (Univ. of Minnesota, USA) and Krystyn J. Van Vliet (MIT/SMART), “Why the dish makes a difference: Quantitative comparison of polystyrene culture surfaces”, Acta Biomaterialia, 9(7), 7354-61 (2013)
- Xinghua Zhang (SMART), Hu Chen (MBI/NUS), Hongxia Fu (MBI/NUS), Patrick S. Doyle (MIT/SMART) and Jie Yan (NUS), "Two distinct overstretched DNA structures revealed by single-molecule thermodynamics measurements", PNAS, 2012, published online on April 24, 2012
- Dai, L. (SMART), Jones, J.J. (MIT), van der Maarel, J.R.C. (NUS), and Doyle, P.S. (MIT/SMART), "A Systematic Study of DNA Conformation in Slitlike Confinement", Soft Matter, 8, 2972-2982 (2012)
- Adam S. Zeiger (MIT/SMART), Felicia C. Loe (SMART), Ran Li (MIT), Michael Raghunath (NUS), and Krystyn J. Van Vliet (MIT/SMART), "Macromolecular crowding directs extracellular matrix organization and mesenchymal stem cell behavior", PLoS, 7(5), e37904 (2012)
- Jing Tang (MIT), Ning Du (SMART) and Patrick Doyle (MIT/SMART), "Compression and self-entanglement of single DNA molecules under uniform electric field", Proceedings of the National Academy of Sciences (PNAS), 108, 16153-16158 (2011)
- Marie-Eve Aubin-Tam, David C. Appleyard, Enrico Ferrari, Valeria Garbin, Oluwatimilehin O. Fadiran, Jacquelyn Kunkel, and Matthew J. Lang, "Adhesion through Single Peptide Aptamers", Journal of Physical Chemistry A, 115: 3657 (2011).
- Bahl N (NUS/SMA), Du R, Winarsih I, Ho B, Tucker-Kellogg L (NUS/MBI), Tidor B (MIT/SMART), Ding JL "Delineation of lipopolysaccharide (LPS)-binding sites on hemoglobin: from in silico predictions to biophysical characterization", J Biol Chem 286: 37793–37803 (2011)
- Huang L, Pan CQ, Li B, Tucker-Kellogg L (NUS/MBI), Tidor B (MIT/SMART), Chen Y, Low BC, "Simulating EGFR-ERK signaling control by scaffold proteins KSR and MP1 reveals differential ligand-sensitivity co-regulated by Cbl-CIN85 and endophilin" PLoS One 6: e22933.
(2011)
- Hyungsuk Lee, Jorge M. Ferrer, Matthew J. Lang, and Roger D. Kamm, "Molecular origin of strain softening in cross-linked F-actin networks", PHYSICAL REVIEW E, 82, 011919 (2010)
- Trahan, D.W. and Doyle, P.S., "DNA collisions with a large, conducting post", Macromolecules, 43, 5424 (2010)
- Hyungsuk Lee, Jorge M. Ferrer, Fumihiko Nakamura, Matthew J. Lang, Roger D. Kamm, "Passive and active microrheology for cross-linked F-actin networks in vitro", Acta Biomaterialia 6, 1207 (2010)
- Tang, J., Trahan, D.W. and Doyle, P.S., "Coil-Stretch Transition of DNA Molecules in Slitlike Confinement", Macromolecules, 43, 3081-3089, 2010.
- Peyman Honarmandi, Hyungsuk Lee, Matthew J. Lang
and Roger D. Kamm, "A microfluidic system with optical laser tweezers to study mechanotransduction and focal adhesion recruitment", Lab on a Chip, 11, 684 (2011)
|
|
- Rajesh Kumar Sharma (SMART/NUS), Patrick S. Doyle (MIT/SMART), Slaven Garaj (NUS), Prevalence of loose and tight knots in DNA investigated by nanopores, APS March Meeting 2018, Los Angeles, CA, USA (Oral Presentation)
- Rajesh Kumar Sharma (SMART/NUS), Patrick S. Doyle (MIT/SMART), Slaven Garaj (NUS), Complex DNA knots observed with solid-state nanopores, BES12SM 2018, Singapore (Merit Award)
- V. Anand Ganesh (SMART), Binu Kundukad (SMART), Dan Cheng (SMART/CENSAM), Sridhar Radhakrishnan (NUS), Seeram Ramakrishna (NUS) and Krystyn J. Van Vliet (MIT), "Novel biofilm resistant nanofibrous membranes for water treatment applications”, 2017, Oral Presentation, 8th IWA Membrane Technology Conference & Exhibition for Water and Wastewater Treatment and Reuse, Singapore
- Sarangapani Sreelatha (SMART), Ajeet kumar Patil (SMART), Rosmin, Matt Lang (Vanderbilt/SMART), Anand Asundi (NTU), "3D/4D Multiscale Imaging in acute lymphoblastic leukemia cells- visualizing dynamics of cell death", ICopen 2017 April 5-7, Singapore Expo.Singapore
- Dai Liang (SMART) and Patrick Doyle (MIT/SMART), "Understanding of knots in polymers by a unified theory", American Physical Society, New Orleans, USA, March 13-17, 2017
- LIU Xiaogang (SMART), TEO Yock Siong (NUS), GU Danning (NUS), Matthew LANG (SMART), “Molecular Design of UV-vis Absorption and Emission Properties in Organic Fluorophores: Enhanced Stokes Shifts with Bright Fluorescence”, The Asian Pacific Conference on Chemistry of Materials 2015, Beijing, China, 17—20 Aug 2015.
- Binu Kundukad (SMART), Chew Su Chuen (SCELSE), Thomas Seviour(SCELSE), Scott Rice(SCELSE), Staffan Kjelleberg(SCELSE) & Patrick S Doyle (SMART, MIT), “Ecomechanics of biofilms”, 10th Annual European Rheology Conference (AERC) 2015, Nantes, France, April 14-17, 2015
- Zhang X. (SMART), “Nonlinear micromechanics of biofilm extracellular polymeric substance revealed by magnetic tweezers” Research Innovation in Infectious and Inflammatory Diseases, 8 July, 2013, Singapore
- Yan J. (NUS)., “Unveiling the mystery of the DNA overstretching transition.” International Conference on Problems of Theoretical Physics (dedicated to the 100th anniversary of outstanding theoretical physicist Alexander Davydov), 8 October 2012; Ukraine
- Yun Soo Kim (SMART), Binu Kundukad (SMART) Abdollah Allahverdi (NTU), Lars Nordenskold (NTU), Patrick S. Doyle (MIT/SMART) and Johan R. C. van der Maarel (NUS). "Using Microrheology to Probe the Gelation of Circular DNA Via Topoisomerase II Clamp Formation and Relation to Anticancer Drugs", ICR 2012-The XVIth International Congress on Rheology, Portugal, August 2012.
- Yun Soo Kim (SMART), Binu Kundukad (SMART) Abdollah Allahverdi (NTU), Lars Nordenskold (NTU), Patrick S. Doyle (MIT/SMART) and Johan R. C. van der Maarel (NUS). “Flow and gelation of DNA by topoisomerase II and some of their targeting cancer therapeutics”, Institut Curie, Paris, France, July '12.
- Binu Kundukad (SMART), Li Yanan (SMART), Cong Piewen (NUS), Yan Jie (NUS), Johan R. C. van der Maarel (NUS), and Patrick S. Doyle (MIT/SMART). “Time dependent stiffening and compaction of DNA by HU proteins” Biological Complex Fluids, Softflow July 2012, Cargese, Corsica Island, France
- Yun Soo Kim (SMART), Binu Kundukad (SMART) Abdollah Allahverdi (NTU), Lars Nordenskold (NTU), Patrick S. Doyle (MIT/SMART) and Johan R. C. van der Maarel (NUS). “Gelation of DNA by topoisomerase II and its targeting anti-cancer drugs”, IUPAC, World Polymer Congress, Blacksburg, USA, June '12
- X. Zhang (SMART), H. Fu, P.S. Doyle (MIT/SMART), and J.Yan (NUS), "Temperature dependence of two distinct DNA overstretching transitions", APS March Meeting, March 2011; Dallas, TX USA.
- Zeiger A, Felicia Loe, Michael Raghunath, & Krystyn J. Van Vliet, "Influence of Altered Local Effective Concentration On Cytoskeletal and ECM Structure in Human Mesenchymal Stem Cells", Biophysical society meeting on Actin & Cytoskeleton (2010)
- Zeiger A, Kotecki M, Maloney J, Herman IM, and Van Vliet KJ , " Pericyte actomyosin-mediated contraction at the cell-material interface can modulate the microvascular niche” –Biophysical society meeting on Actin & Cytoskeleton (2010)
- John M. Maloney, Ranjani Krishnan, Adam Zeiger, and Krystyn J. Van Vliet, “Chemomechanics of Cell-Material Interaction Kinetics Over Large Ranges of Length and Time Scales, “Materials Research Society Fall Meeting, Boston, MA, Dec. 1, 2009.
|
|
 |